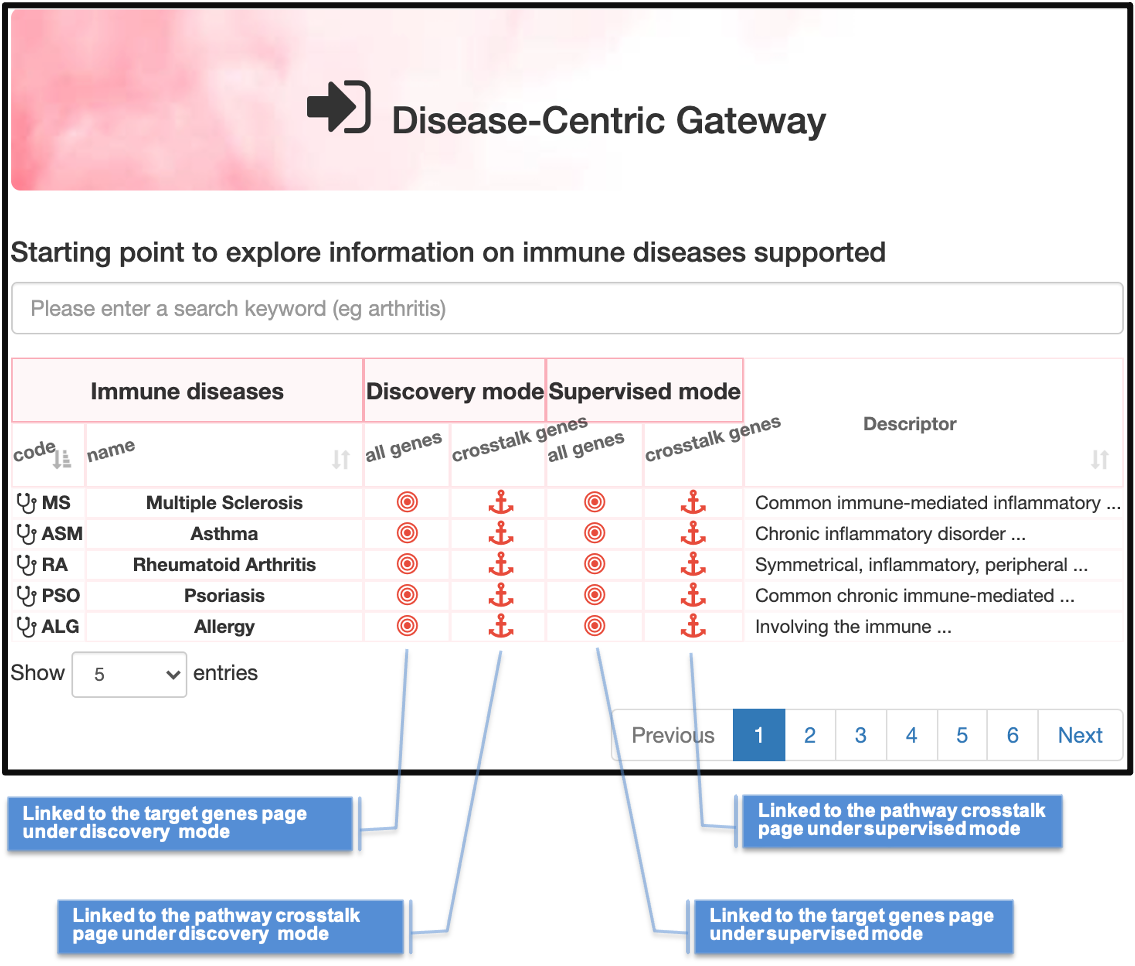
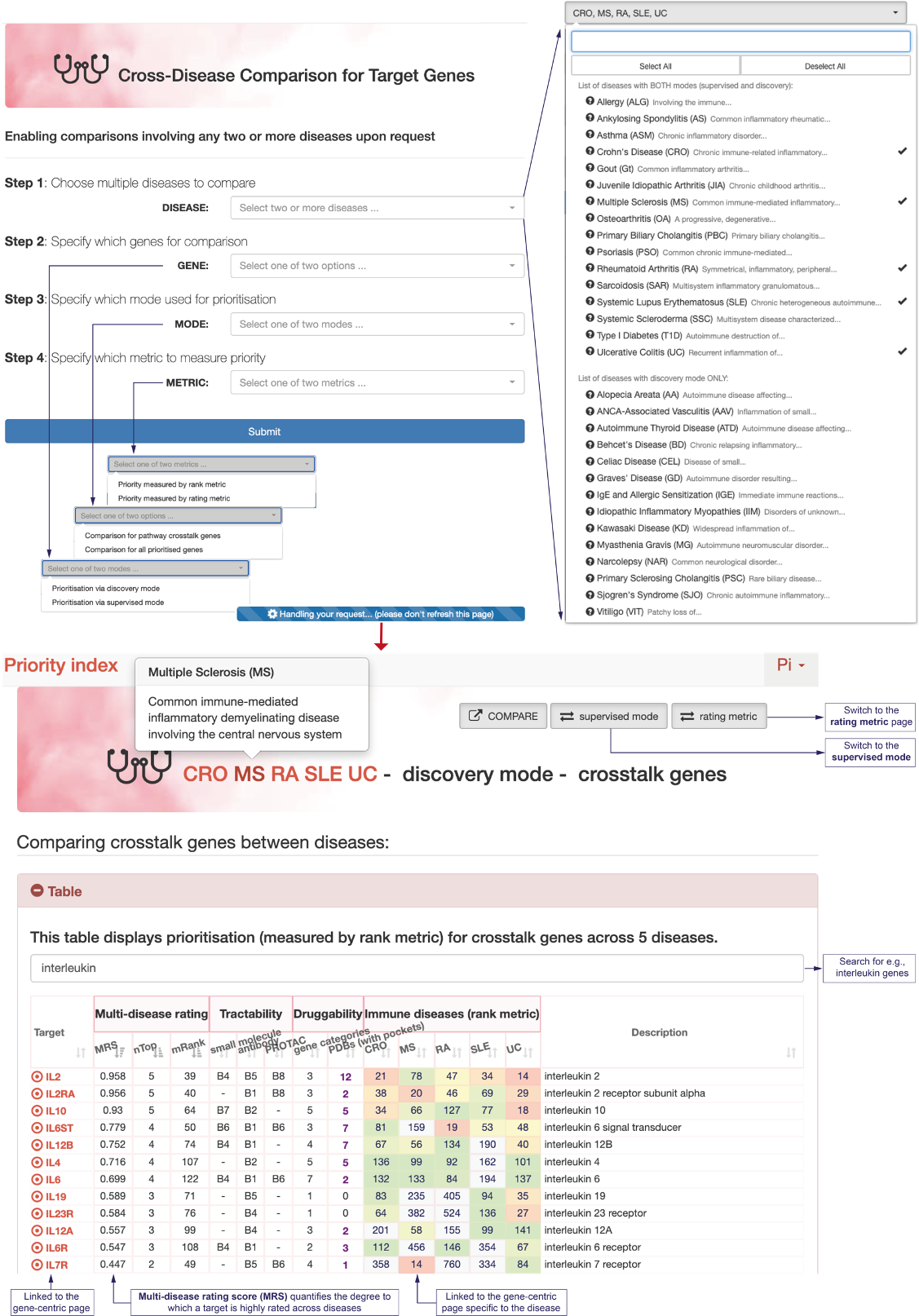
FRONTPAGE
Landing frontpage of the Pi web, with faceted search returning diseases, genes (linked page illustrated) and structures (3D view illustrated)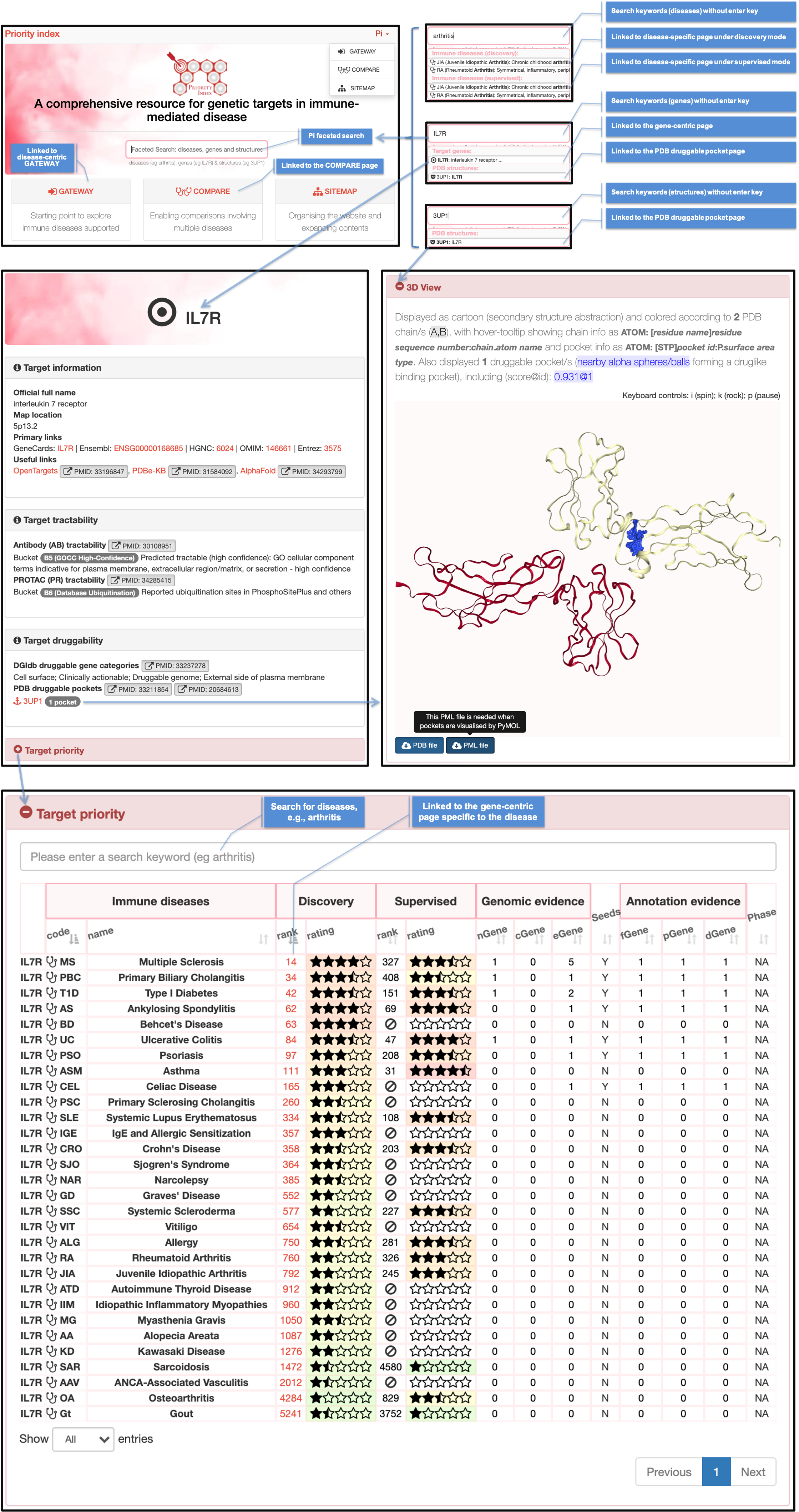
|
|
|
DISEASE-SPECIFIC PAGES
Illustrating 1) the disease-specific page on target genes; and 2) the gene-centric page specific to the disease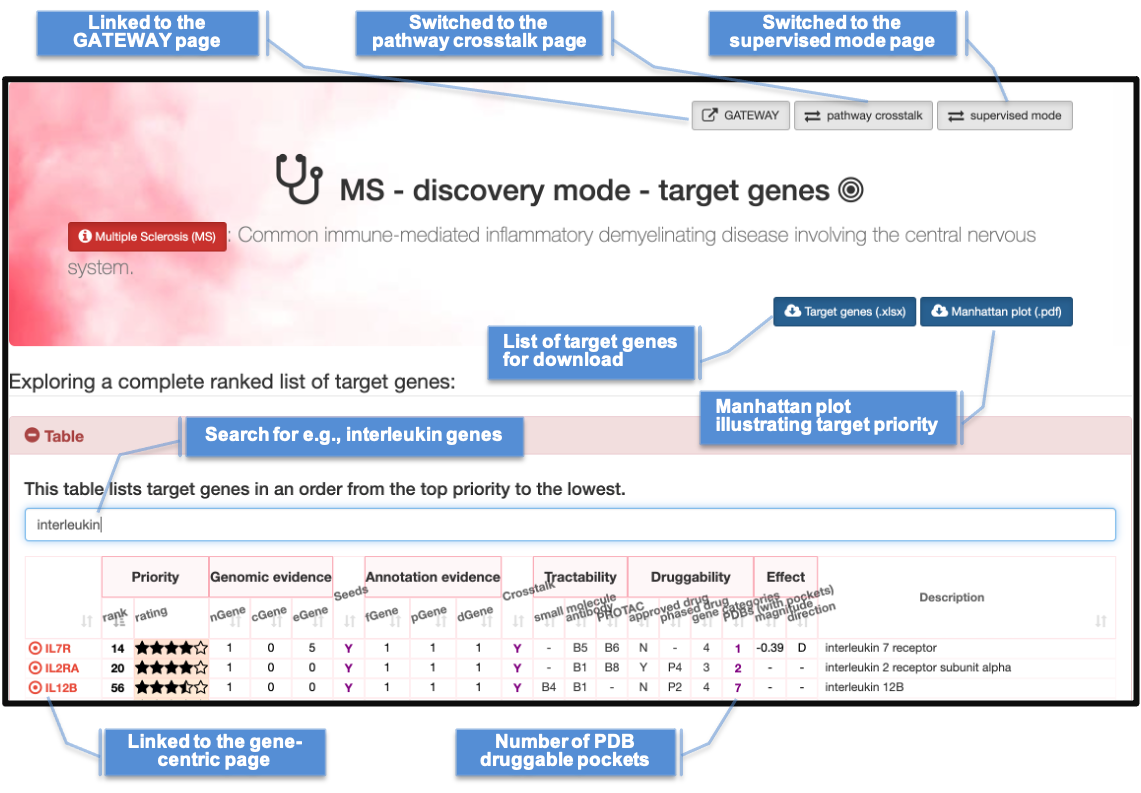
|
|
Disease-specific page on target genes |
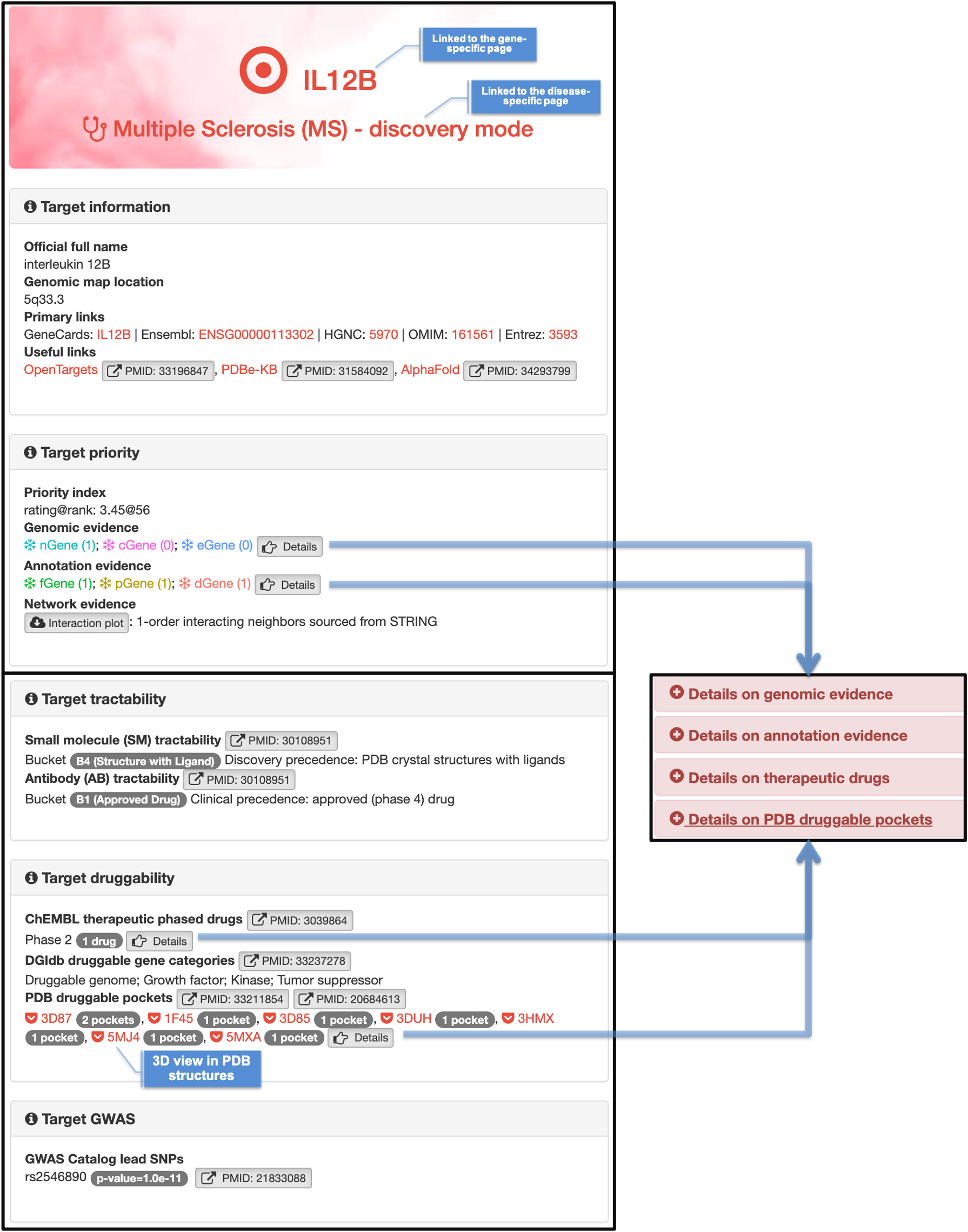
|
|
Gene-centric page specific to the disease |
pi_database_202108.sql.gz containing 7 tables designed to store database contents.
pi_bucket
Containing info about target tractability (organised as buckets)
mysql> DESC pi_bucket;
+-------------+----------------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+-------------+----------------------+------+-----+---------+-------+
| gene | varchar(20) | NO | PRI | NULL | |
| modality | enum('AB','PR','SM') | NO | PRI | NULL | |
| bucket | varchar(2) | NO | PRI | | |
| name | varchar(50) | NO | | NULL | |
| description | varchar(200) | NO | | NULL | |
+-------------+----------------------+------+-----+---------+-------+
- The gene column is the gene symbol
- The modality column can be one of three drug modalities including antibody (AB), small molecule (SM) and PROTAC (PR)
- The bucket column is the bucket identifier starting with the letter 'B' followed by an integer
- The name column is the bucket name
- The description column is the description of buckets detailing the tractability evidence
pi_category
Containing info about gene druggable categories sourced from DGIdb
mysql> DESC pi_category;
+----------+-------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+----------+-------------+------+-----+---------+-------+
| gene | varchar(20) | NO | PRI | NULL | |
| category | varchar(50) | NO | PRI | | |
+----------+-------------+------+-----+---------+-------+
- The gene column is the gene symbol
- The category column shows gene druggable categories
pi_disease
Containing info about immune-mediated diseases
mysql> DESC pi_disease;
+------------+---------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+------------+---------------+------+-----+---------+-------+
| disease | varchar(3) | NO | PRI | NULL | |
| name | varchar(30) | NO | | NULL | |
| descriptor | varchar(1000) | NO | | NULL | |
+------------+---------------+------+-----+---------+-------+
- The disease column is the 2- or 3-letter disease code
- The name column is the disease name
- The descriptor column is the short descriptor for disease
pi_drug
Containing info about drug therapeutics sourced from ChEMBL
mysql> DESC pi_drug;
+---------------------+--------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+---------------------+--------------+------+-----+---------+-------+
| disease | varchar(5) | NO | PRI | | |
| gene | varchar(20) | NO | PRI | NULL | |
| max_phase | int unsigned | NO | PRI | 0 | |
| drug | varchar(255) | NO | PRI | | |
| mechanism_of_action | varchar(250) | NO | PRI | | |
| action_type | varchar(50) | YES | | NULL | |
| source | varchar(500) | NO | | NULL | |
+---------------------+--------------+------+-----+---------+-------+
- The disease column is the 2- or 3-letter disease code
- The gene column is the gene symbol
- The max_phase column is the maximum phase of drug development reached
- The drug column is the drug name
- The mechanism_of_action column is the mechanism of action for drug targeting
- The action_type column is the action type for drug targeting
- The source column shows the primary sources for drug targeting
pi_genomic
Containing info about genomic evidence
mysql> DESC pi_genomic;
+-----------+-----------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+-----------+-----------------+------+-----+---------+-------+
| disease | varchar(5) | NO | PRI | | |
| gene | varchar(20) | NO | PRI | NULL | |
| gene_type | varchar(5) | NO | | NULL | |
| cell_type | varchar(100) | NO | PRI | NULL | |
| snp | varchar(15) | NO | PRI | NULL | |
| pvalue | double unsigned | YES | | NULL | |
+-----------+-----------------+------+-----+---------+-------+
- The disease column is the 2- or 3-letter disease code
- The gene column is the gene symbol
- The gene_type column is the type of genomic seed genes
- The cell_type column lists the cell types
- The snp column is the dbSNP identifier
- The pvalue column is the p-values detected by GWAS
pi_pdb
Containing info about druggable pockets predicted based on known PDB structures
mysql> DESC pi_pdb;
+-----------+---------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+-----------+---------------+------+-----+---------+-------+
| gene | varchar(20) | NO | PRI | NULL | |
| uniprot | varchar(10) | NO | PRI | NULL | |
| pdb_chain | varchar(15) | NO | PRI | NULL | |
| pdb | varchar(5) | NO | | NULL | |
| chain | varchar(10) | NO | | NULL | |
| pocket | enum('Y','N') | NO | | NULL | |
+-----------+---------------+------+-----+---------+-------+
- The gene column is the gene symbol
- The uniprot column is the UniProt identifier
- The pdb_chain column shows the PDB chain in the structure
- The pdb column is the PDB code
- The chain column shows which PDB chain
- The pocket column specifies whether or not the PDB chain contains the druggable pockets
pi_priority
Containing info about target priority
mysql> DESC pi_priority;
+--------------------+--------------------+------+-----+---------+-------+
| Field | Type | Null | Key | Default | Extra |
+--------------------+--------------------+------+-----+---------+-------+
| disease | varchar(5) | NO | PRI | | |
| gene | varchar(20) | NO | PRI | NULL | |
| rank | int unsigned | YES | | NULL | |
| rating | double | YES | | NULL | |
| nGene | mediumint unsigned | NO | | 0 | |
| cGene | mediumint unsigned | NO | | 0 | |
| eGene | mediumint unsigned | NO | | 0 | |
| seed | enum('Y','N') | NO | | NULL | |
| dGene | mediumint unsigned | NO | | 0 | |
| pGene | mediumint unsigned | NO | | 0 | |
| fGene | mediumint unsigned | NO | | 0 | |
| crosstalk | enum('Y','N') | NO | | NULL | |
| phased | varchar(2) | YES | | NULL | |
| druggable_category | mediumint unsigned | NO | | 0 | |
| pdb_pocket | mediumint unsigned | NO | | 0 | |
| magnitude | double | YES | | NULL | |
| direction | enum('I','D','-') | NO | | NULL | |
| description | varchar(1000) | NO | | NULL | |
+--------------------+--------------------+------+-----+---------+-------+
- The disease column is the 2- or 3-letter disease code
- The gene column is the gene symbol
- The rank column shows priority rank
- The rating column shows priority rating (scored 0-5)
- The nGene column specifies whether or not the nearby gene
- The cGene column shows how many conformation genes (cell types) are identified
- The eGene column shows how many eQTL genes (cell types) are identified
- The seed column specifies whether or not it is a genomic seed gene (nGene, cGene or eGene)
- The dGene column specifies whether or not the disease gene
- The pGene column specifies whether or not the phenotype gene
- The fGene column specifies whether or not the function gene
- The crosstalk column specifies whether or not the pathway crosstalk gene
- The phased column shows the drug development phase information targeting the gene, namely, preclinical phases 0-3 (P0-P3), phase 4 (P4; approved), otherwise '-'
- The druggable_category column shows the number of gene druggable categories
- The pdb_pocket column shows the number of druggable pockets based on PDB structures
- The magnitude column specifies the estimates of effect size (magnitude) on disease
- The direction column specifies the estimates of effect direction on disease
- The description column is the gene description
USAGE
Restoring the database
mysql -u root -p -e "create database pi;"
gunzip < pi_database_202108.sql.gz | mysql -u root -p pi
Showing tables
mysql> USE pi; mysql> SHOW tables; +-------------+ | pi_bucket | | pi_category | | pi_disease | | pi_drug | | pi_genomic | | pi_pdb | | pi_priority | +-------------+
Extracting pathways crosstalk genes in Multiple Sclerosis (MS)
mysql> SELECT gene,rating,description FROM pi_priority WHERE disease='MS' and crosstalk='Y'; +----------+------------------+------------------------------------------------------------+ | gene | rating | description | +----------+------------------+------------------------------------------------------------+ | CD3D | 3.35080030156153 | CD3d molecule | | CD3E | 3.42984078279532 | CD3e molecule | | CD3G | 3.36283894499324 | CD3g molecule | | CD4 | 3.84211138613646 | CD4 molecule | | CD40 | 4.09069218691624 | CD40 molecule | | CD80 | 3.51309447797322 | CD80 molecule | | CD86 | 3.86358361572689 | CD86 molecule | | CD8A | 3.7713984905519 | CD8a molecule | | CDK4 | 3.36314860797353 | cyclin dependent kinase 4 | | GH1 | 3.28003935980419 | growth hormone 1 | | IFNG | 3.35178195680181 | interferon gamma | | IFNGR1 | 3.68129957906975 | interferon gamma receptor 1 | | IL10 | 3.39301077350483 | interleukin 10 | | IL12A | 3.44597274350518 | interleukin 12A | | IL12B | 3.45064200047236 | interleukin 12B | | IL2 | 3.26387267975032 | interleukin 2 | | IL2RA | 3.77737122651625 | interleukin 2 receptor subunit alpha | | IL7R | 3.89020505777676 | interleukin 7 receptor | | IRF1 | 3.55134023208536 | interferon regulatory factor 1 | | IRF3 | 3.45328702733898 | interferon regulatory factor 3 | | IRF5 | 3.52267944150431 | interferon regulatory factor 5 | | IRF9 | 3.55064925773335 | interferon regulatory factor 9 | | LCK | 3.18952838424379 | LCK proto-oncogene, Src family tyrosine kinase | | MAPK1 | 4.25349978965674 | mitogen-activated protein kinase 1 | | MYC | 3.38620467690507 | MYC proto-oncogene, bHLH transcription factor | | NFKB1 | 3.73865859299998 | nuclear factor kappa B subunit 1 | | NFKBIA | 3.47571622561559 | NFKB inhibitor alpha | | SOCS1 | 3.70599921146337 | suppressor of cytokine signaling 1 | | STAT3 | 3.40946436915169 | signal transducer and activator of transcription 3 | | TNFRSF1A | 3.62110604866511 | TNF receptor superfamily member 1A | | TRAF2 | 2.95384907354841 | TNF receptor associated factor 2 | | TRAF3 | 3.2668223879653 | TNF receptor associated factor 3 | | TYK2 | 3.5565594737874 | tyrosine kinase 2 | | VCAM1 | 3.46078655932035 | vascular cell adhesion molecule 1 | | ZAP70 | 3.15408662162023 | zeta chain of T cell receptor associated protein kinase 70 | +----------+------------------+------------------------------------------------------------+